Intelligent Digital Inline Holographic Micrograph (DIHM) Cell-Enhancement and Characterization
Intelligent Digital Inline Holographic Micrograph (DIHM) Cell-Enhancement and Characterization
Project Intro
Developing a novel and efficient neural model for detecting and classifying cells such as RBC, WBC, and cancer cells, etc.(cellular pathology) from DIH(Digital Inline Holographic) microscopy images
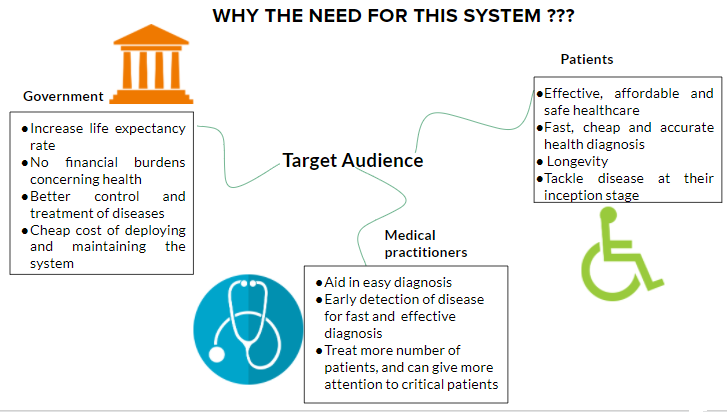
Implementation on a resource-constrained device to develop a cheap, reliable and portable point-of-care testing facility for diagnosis of pathological diseases, especially for usage in rural areas
Highlights
- Segment cell-lines in DIH micrograph; performs signal enhancement using CNN-based autoencoder, followed by the cell-line characterization
- ROC-AUC: >0.98 for RBC, WBC, and microbeads; >0.88 for cancer cells HepG2 and MCF7
- Easy accommodation of newer cell-lines. Python, TensorFlow, OpenCV
- Preliminary work published at the 8th IEEE ICHI
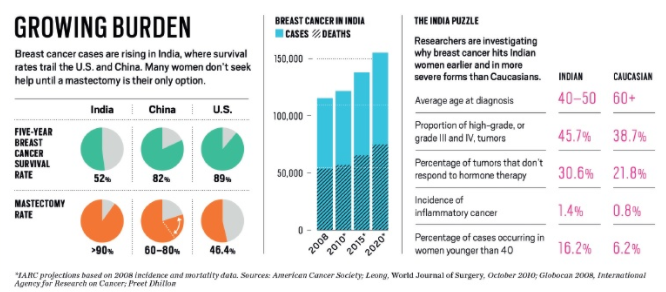
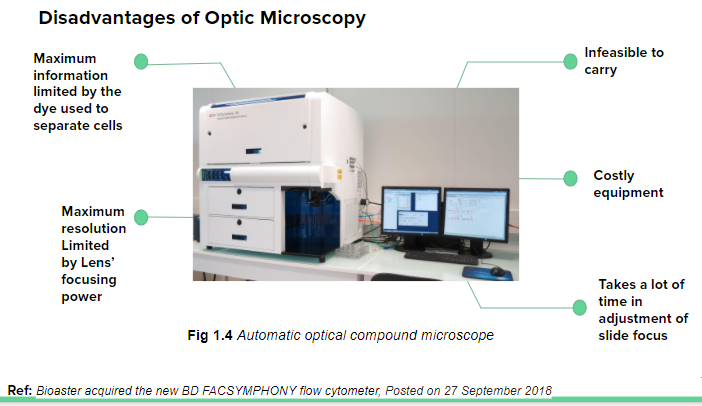
Aim
The aim of the research is to create a
- Cheap,
- Reliable,
- Adaptable,
- Portable, and
- Intelligent real-time point-of-care testing facility that could be used in resource-constrained environments, especially such as those in a rural setting.
Dataset
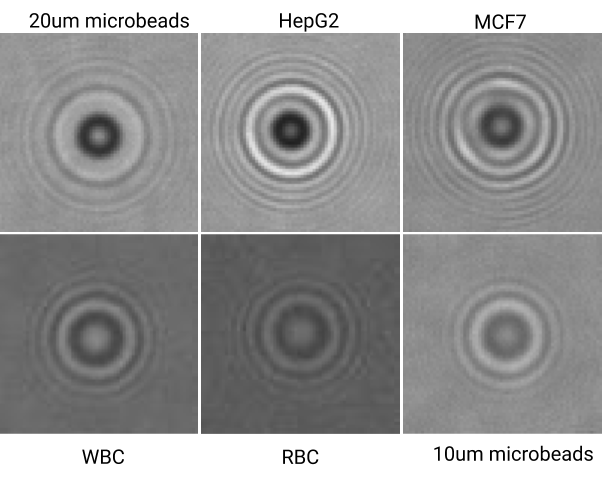
Current Progress
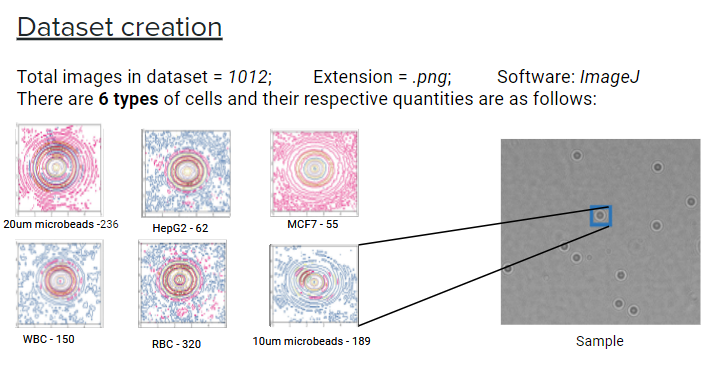
The colored cell-lines is the result of EDA on the statistical properties and pixel intensity in the images
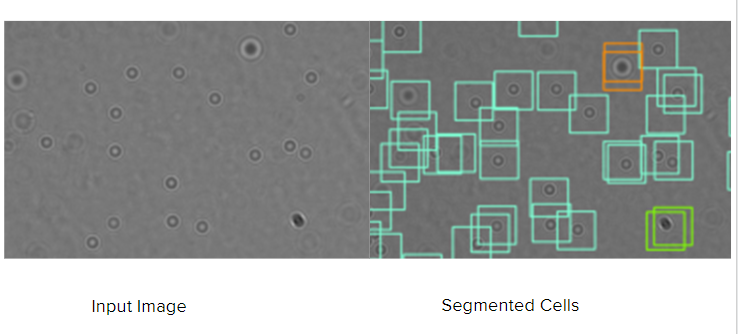
Segmentation Results
CNN Model Performance
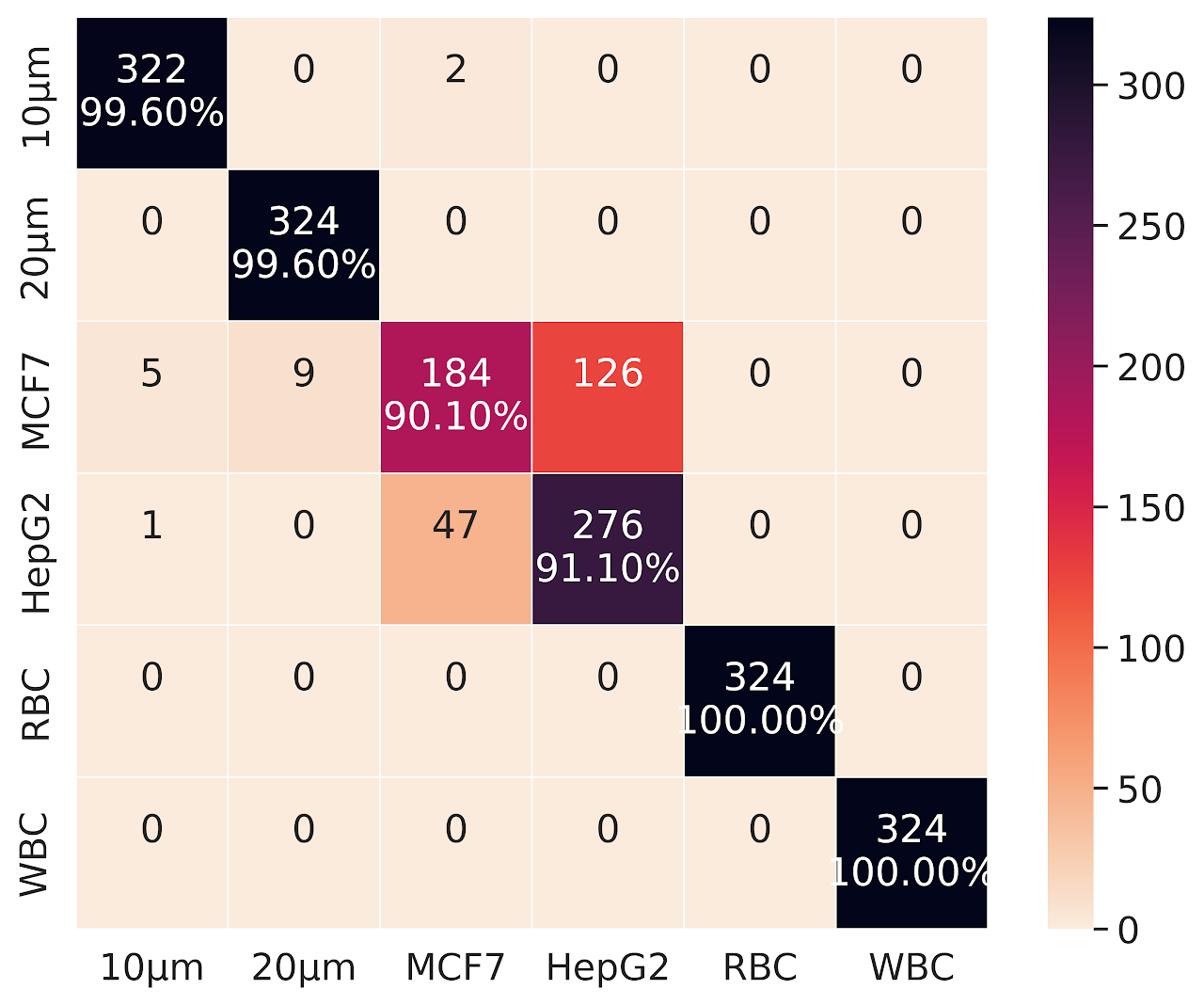
The recognition performance of the CNN model on the augmented dataset
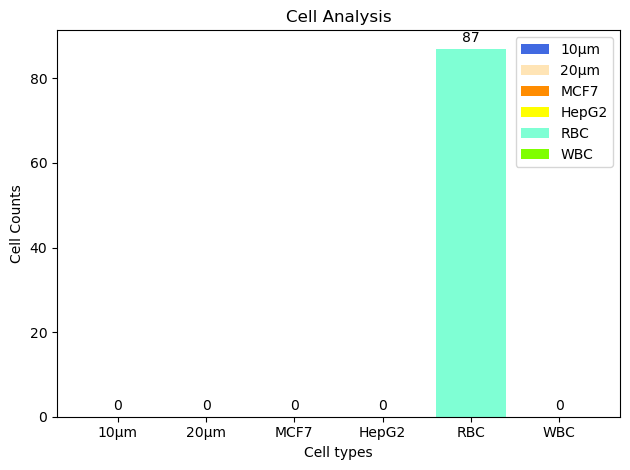
Final Cell Counts for the Input Micrograph